Cimwch.com
An article which appeared in "Crustaceana" 77 (3): 371-376
A Potential Method For Discriminating Between Tissue From The European Lobster (Homarus gammarus) And The American Lobster (H. americanus).
GRETA HUGHES and ANDY R. BEAUMONT
School of Ocean Sciences, University of Wales, Bangor, Menai Bridge, Gwynedd, LL59 5AB, United Kingdom.
INTRODUCTION
The American lobster, Homarus americanus (L., 1758) is native to waters off the north-eastern coast of the U.S.A. and Canada and large numbers of live H. americanus are imported into Europe where they are held in licensed storage tanks before sale. Reports occasionally appear of American lobsters being caught from the wild in Europe, most recently in Norway, probably as a result of accidental release from holding tanks, or alternatively having been jettisoned from cruise liners (Laing, 2002). European countries are becoming increasingly concerned about the potential consequences of accidental releases of live H. americanus in European waters where they could interbreed with the European lobster, Homarus gammarus (L., 1758). Hybrid offspring from these two species have been produced in the laboratory and while males are infertile, females may produce fertile eggs if back-crossed with wild stock (Hedgecock et al., 1978) so there is the potential for considerable genetic impact. In addition, the blood disease, gaffkemia, caused by the bacterium, Aerococcus viridans, is endemic in wild H. americanus and causes heavy mortality in storage ponds. The pathogen can live and multiply outside the lobster in the slime on transportation boxes and tanks and has also been isolated from mud of tidal ponds and from seawater several miles from infected storage ponds (Goggins & Hurst, 1960). At this time there is no evidence that the disease is present in H. gammarus populations.
There is a characteristic ventral tooth on the rostrum of H. americanus that is absent from H gammarus (Holthuis, 1991), enabling the two species to be quite easily identified morphologically as long as the rostrum is present. Nevertheless, H. americanus claws or other tissues sold separately cannot be distinguished from similar H. gammarus tissues and therefore a simple species-specific genetic marker would be valuable for forensic investigations.
Inter-species genetic differences between Homarus gammarus and H. americanus have been identified by analysis of the mitochondrial 16s ribosomal RNA gene sequence and by the use of three microsatellite loci (Kornfield et al., 1995; Tam & Kornfield, 1996). In addition there has been an extensive recent study on the population genetics of lobsters using microsatellites (Ferguson, 2002; Hughes, 2003) The random amplified polymorphic DNA (RAPD) (Williams et al., 1990) method has detected genetic variation in H. gammarus (Ulrich et al., 2001), H. americanus (Harding et al., 1997) and the penaeid prawns, Penaeus vannamei (Garcia et al., 1994) and P. monodon (Garcia & Benzie, 1995; Tassanakajon et al., 1997). Here we report on species-specific RAPD markers for distinguishing between DNA from H. americanus and H. gammarus tissues.
MATERIALS AND METHODS
Pereiopods were removed from 21 Canadian Homarus americanus held in tanks by Smith Sager Ltd., Manchester, in October, 1998. Between 1992 and 1997, pereiopods were removed from approximately equal numbers (+15) live male and female H. gammarus from commercial holding tanks belonging to individual fishermen or shellfish merchants from locations around the coasts of the U.K., Ireland, and Norway (table I).
Table I
Locations, sample size and date of tissue collection of 10 samples of Homarus gammarus (L.) and one H. americanus (L.) sample used for RAPD analysis. The Ireland sample was first imported to Covine Fisheries, Weymouth and collected from there.
|
Location |
Sample size |
Date |
|
Bridlington, Yorkshire, England |
30 |
June, 1992 |
|
West coast of Ireland, Eire |
30 |
June, 1993 |
|
Johnshaven, Grampian, Scotland |
30 |
June, 1994 |
|
Llyn Peninsula, Gwynedd, North Wales |
30 |
August, 1997 |
|
Mevagissey, Cornwall, England |
30 |
June, 1993 |
|
East Runton, Norfolk, England |
30 |
June, 1992 |
|
Austevoll, Norway |
30 |
July, 1998 |
|
Isles of Scilly, England |
30 |
June, 1993 |
|
North Uist, Outer Hebrides, Scotland |
30 |
June, 1994 |
|
Weymouth, Dorset, England |
30 |
June, 1993 |
|
Homarus americanus, Canada |
21 |
October, 1998 |
Approximately 5 mm3 of pereiopod tissue was placed in a 1.5 ml microtube and plunged into liquid nitrogen within 5 minutes of dissection and transferred to a –70°C deep freeze within 16 days. DNA was extracted using SDS and proteinase K followed by standard phenol chloroform extraction (Sambrook et al., 1989; Garcia & Benzie, 1995). The DNA pellet was finally suspended in 25 µl TE buffer and stored at 4°C until required.
Initial PCR trials were conducted with 20 different RAPD 10bp primers (Operon Technologies, Almeda, CA’s Kit-R) to screen for primers that produced repeatable banding patterns. Following optimization, PCR reactions were performed in a total volume of 25 µl containing 17.2 µl pure water, 2.5 µl thermo buffer, 0.2 µl dNTPs (final molarity 1 mM), 2 µl MgCl2 (final molarity 25 mM) 0.1 µl (0.5 units) Taq polymerase, 0.5 µl primer (final molarity 0.3 µM), and 2.5 µl of a 1 : 10 dilution of template DNA. PCR amplification was optimized to: 1 min. of initial denaturation at 94.5°C, then 10 cycles of 40 seconds at 94.5°C, 1 min. at 33°C, and 2 min. at 72°C, followed by 35 cycles of 30 seconds at 94.5°C, 1 min. at 35°C, and 2 min. at 72°C with a final extension of 5 minutes at 72°C before cooling to 4°C. A negative control with pure water substituted for template DNA was included in every PCR run to identify potential artefact bands.
PCR products were separated on 2.5% agarose gel, stained with ethidium bromide and visualized under UV light. Bands were scored from photographs taken of each gel.
RESULTS
Primer sequences suitable as species-specific markers are given in table II.
Table II
RAPD primer sequences useful for discrimination between the DNA of Homarus americanus (L.) and H. gammarus (L.).
|
Primer |
Sequence (5’–3’) |
Species-specific product in H. americanus |
Species-specific product in H. gammarus |
|
OPR-01 |
TGCGGGTCCT |
410 bp* |
|
|
OPR-08 |
CCCGTTGCCT |
510 bp |
|
|
OPR-20 |
ACGGCAAGGA |
No product |
20 bands (375-1500 bp) |
*Similar sized fragment found in two H. gammarus individuals from Norfolk.
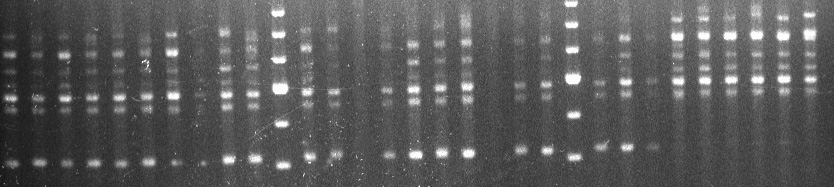
Primer OPR-01 amplified a 410 bp band from all 20 Homarus americanus samples and from 2 H. gammarus individuals, both from Norfolk (fig. 1). A second primer, OPR-08, produced a 510 bp fragment that was specific to H. americanus and present in 18 out of 20 samples.
H. americanus H. gammarus
Marker Marker
410bp 
Fig.1, Bands amplified by primer OPR 1 for Homarus americanus (L. 1758) and Homarus gammarus (L. 1758); OPR410 is absent in H. gammarus.
In contrast, primer OPR-20 failed to amplify any PCR products from H. americanus DNA but amplified up to 20 bands (ranging from 375 to 1500 bp) from H. gammarus DNA. Full data on the RAPD PCR products from H. gammarus are given elsewhere (Hughes, 2000).
DISCUSSION
The results indicate that DNA from Homarus spp. can be distinguished using the RAPD method. The safest test would be to screen PCR products from all three primers (OPR-01, OPR-08, and OPR-20) because the expected OPR-01410 and OPR-08510 PCR products from H. americanus were very occasionally difficult to detect. The RAPD methodology has suffered from legitimate criticism because of the occasional unreliability and unrepeatability of PCR products (Patwary et al., 1994). With such a low annealing temperature during PCR (in this case 33-35 oC) it is expected that some primer mismatches are likely to occur which will result in weak, or non-repeatable bands. This is a particular problem when using RAPDs for population genetic studies but is less of a concern when looking at differences between species based on the presence of a clear PCR product in one species and its complete absence in the other species. The RAPD species-specific bands for Homarus spp. are generally strong and clear and there is a large sample size (approximately 300: Hughes, 2000) of H. gammarus from a number of locations confirming the almost complete absence of the OPR-01410 and the complete absence of the OPR-08510 bands in this species. Due to the limited resolution of a 2.5% agarose gel, it was not possible to confirm that the OPR-01410 band seen in the two Norfolk H. gammarus was exactly the same size as that in H. americanus individuals but this could be confirmed by using a thin layer polyacrylamide gel or removing the fragment from an agarose gel and sequencing it (Hadrys et al.s 1992).
Other methods can distinguish between Homarus spp. DNA (Kornfield et al., 1995; Tam & Kornfield, 1996) but they involve the use of a DNA sequencer, or radio-labelling methodology. In contrast, the RAPD method requires only a PCR machine and simple agarose gel electrophoresis.
Further development of the methodology, such as multiplex PCR, and the use of a much larger sample of H. americanus is required before results from these RAPD products could be used as forensic evidence, but these tests have the potential to be a rapid, cheap, reliable, and effective species-specific marker.
ACKNOWLEDGEMENTS
We are grateful to Grö van der Meeren (Research Institute, Austevol, Norway) for providing the European lobsters from Norway.
REFERENCES
FERGUSON, A. F., (2002) Genetic diversity in the European lobster (Homarus gammarus): population structure and impact on stock enhancement (GEL). FAIR CT984266 Final Report. Available at: http://www.quab.ac.uk/bb/prodohl/GEL.gel.html
GARCIA, D.K. & J.A.H. BENZIE, (1995). RAPD markers of potential use in penaeid prawn (Penaeus monodon) breeding programs. Aquaculture, 130, 137-144.
GARCIA, D.K., M.A. FAGGART, L. RHOADES & A.A. ALCIVAR-WARREN, (1994). Genetic diversity of cultured Panaeus vannamei shrimps using three molecular genetic techniques. Molecular Marine Biology and Biotechnology, 3, 270-280.
GOGGINS, P.L. & J.W. HURST, (1960). Progress report on lobster gaffkyaremia (Red Tail): (Unpublished Report of the Department of Sea and Shore Fisheries, Augusta, Maine).
HADRYS, S.H., M. BALICK & B. SCHIERWATER, (1992). Applications of random amplified polymorphic DNA (RAPD) in molecular ecology. Molecular Ecology, 1, 55-63.
HARDING, G.C., E.L. KENCHINGTON, C.J. BIRD, D.S. PEZZACK & D.C. LANDRY, (1997). Genetic relationships among subpopulations of the American lobster (Homarus americanus) as revealed by random amplified polymorphic DNA. Canadian Journal of Fisheries and Aquatic Sciences, 54, 1762-1771.
HEDGECOCK, D., W.L. MOFFET, W. BORGESON & K. NELSON, (1978).
Progress and problems in lobster broodstock development. Proceedings of the Annual Meeting, World Mariculture Society, 9, 947-506.
HOLTHIUS, L.B., (1991). FAO species catalogue. Marine lobsters of the world. An annotated and illustrated catalogue of species of interest to fisheries known to date. FAO Fisheries Synopsis, 125, 13.
HUGHES, G., (2000). Population structure and genetics of the European lobster, Homarus gammarus: 1-236. (Ph.D. Thesis, University of Wales).
HUGHES, M. F., (2003). A comparative microsatellite study of the population genetic structure and mating system of the European lobster (Homarus gammarus) and of the Norway lobster (Nephrops norvegicus): 1-250. (Ph.D. Thesis, Queens University of Belfast, Belfast).
KORNFIELD, I., A.B. WILLIAMS, & R.S. STENECK, (1995). Assignment of Homarus capensis (Herbst, 1792), the Cape lobster of South Africa, to the new genus, Homarinus (Decapoda:Nephropidae). Fisheries Bulletin, U.S., 93, 97-102.
LAING, I. (2002) American lobsters – over here? Shellfish News, 14: 20-22.
PATWARY, M. U., E.L. KENCHINGTON, C.J. BIRD, & E. ZOUROS, (1994). The use of random amplified polymorphic DNA markers in genetic studies of the sea scallop Placopecten magellanicus (Gmelin, 1791). Journ. Shellfish Res., 13: 547-553.
SAMBROOK, J., E.F. FRITSCH & T. MANIATIS, 1989. Molecular cloning: a laboratory manual (2nd ed.): (Cold Spring Harbor Laboratory Press, New York).
TASSANAKAJON, A., S. PONGSOMBOON, V. RIMPHANITCHAYAKIT, P. JARAYABHAND & V. BOONSAENG, (1997) Random amplified polymorphic DNA (RAPD) markers for determination of genetic variation in wild populations of the black tiger prawn (Penaeus monodon) in Thailand. Molecular Marine Biology and Biotechnology, 6: 110-115.
TAM, Y. K. & I. KORNFIELD, (1996) Characterization of microsatellite markers in Homarus (Crustacea: Decapoda). Molecular Marine Biology and Biotechnology, 5: 230-238.
ULRICH, I., J. MULLER, C. SCHUTT & F. BUCHHOLZ, (2001) A study of population genetics in the European lobster, Homarus gammarus (Decapoda, Nephropidae). Crustaceana, 74: 825-837.
WILLIAMS, J. G. K., A.R. KUBELIK, K.J. LIVAK, J.A. RAFALSKI & S. TINGEY, (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18: 6531-6535.
Copyright © Cimwch.com 2004